General Information
-
DRAMP ID
- DRAMP00249
-
Peptide Name
- Pln149 (Plantaricin 149; Bacteriocin; Derivatives: Pln149a)
-
Source
- Lactobacillus plantarum NRIC 149 (Gram-positive bacteria)
-
Family
- Not found
-
Gene
- Not found
-
Sequence
- YSLQMGATAIKQVKKLFKKKGG
-
Sequence Length
- 22
-
UniProt Entry
- No entry found
-
Protein Existence
- Not found
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Antifungal
-
Target Organism
- No MICs found in DRAMP database
-
Hemolytic Activity
-
- No hemolysis information or data found in the reference(s) presented in this entry
-
Cytotoxicity
-
- Not included yet
-
Binding Target
- Cell membrance
Structure Information
-
Linear/Cyclic
- Not included yet
-
N-terminal Modification
- Not included yet
-
C-terminal Modification
- Not included yet
-
Nonterminal Modifications and Unusual Amino Acids
- Not included yet
-
Stereochemistry
- Not included yet
-
Structure
- Alpha helix
-
Structure Description
- Not found
-
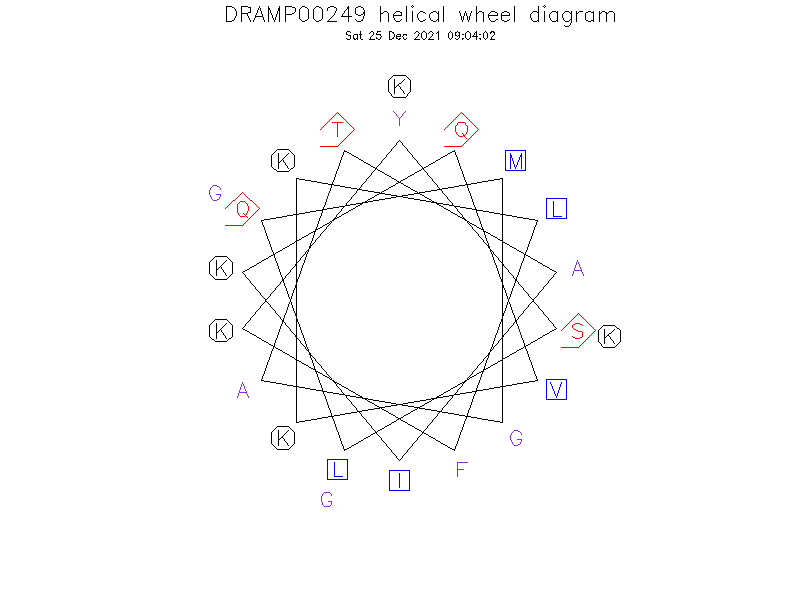
Helical Wheel Diagram
-
PDB ID
- None
-
Predicted Structure
- There is no predicted structure for DRAMP00249.
Physicochemical Information
-
Formula
- C111H190N30O28S
Absent Amino Acids
- CDEHNPRW
Common Amino Acids
- K
Mass
- 2424.97
PI
- 10.4
Basic Residues
- 6
Acidic Residues
- 0
Hydrophobic Residues
- 7
Net Charge
- +6
-
Boman Index
- -19.92
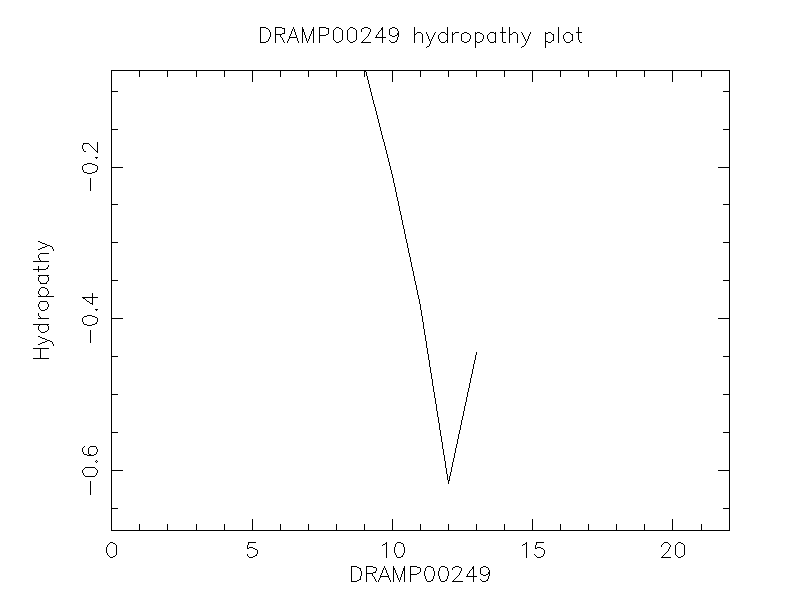
Hydrophobicity
- -0.446
Aliphatic Index
- 75.45
Half Life
-
- Mammalian:2.8 hour
- Yeast:10 min
- E.coli:2 min
Extinction Coefficient Cystines
- 1490
Absorbance 280nm
- 70.95
Polar Residues
- 6
DRAMP00249
Comments Information
Comment
- No comments found on DRAMP database
Literature Information
- ·Literature 1
-
Title
- Plantaricin-149, a bacteriocin produced by Lactobacillus plantarum NRIC 149.
-
Pubmed ID
- 17266050
-
Reference
- J. Fermentation Bioeng. 1994;77(3):277-282.
-
Author
- Kato T, Matsuda T, Ogawa E, Ogawa H, Kato H, Doi U.