General Information
-
DRAMP ID
- DRAMP03532
-
Peptide Name
- Moricin-1 (Insects, animals)
-
Source
- Bombyx mori (Silk moth)
-
Family
- Belongs to the moricin family
-
Gene
- MOR1
-
Sequence
- AKIPIKAIKTVGKAVGKGLRAINIASTANDVFNFLKPKKRKH
-
Sequence Length
- 42
-
UniProt Entry
- P82818
-
Protein Existence
- Protein level
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram+, Anti-Gram-
-
Target Organism
-
- Gram-negative bacteria: Escherichia coli JM109 (MIC=0.31 µM), Acinetobacter sp. NISL B-4653 (MIC=0.27 µM), Pseudomonas fluorescens IAM1179 (MIC=0.53 µM), Pseudomonas aeruginosa IAM15140 (MIC=0.81 µM);
- Gram-positive bacteria: Bacillus subtilis IAM1107 (MIC=0.19 µM), Bacillus megaterium IAM1030 (MI=0.09 µM), Bacillus cereus IFO3457 (MIC=0.38 µM), Staphylococcus aureus ATCC6538P (MIC=0.21 µM), Staphylococcus aureus ATCC6538P (MIC=0.22 µM), Staphylococcus aureus IFO3083 (MIC=0.46 µM), Staphylococcus xylosus IAM1312 (MIC=0.27 µM), Staphylococcus epidermidis IFO12993 (MIC=0.18 µM), Streptococcus pyogenes ATCC21547 (MIC=0.25 µM).
-
Hemolytic Activity
-
- No hemolysis information or data found in the reference(s) presented in this entry
-
Cytotoxicity
-
- Not included yet
-
Binding Target
- Not found
Structure Information
-
Linear/Cyclic
- Linear
-
N-terminal Modification
- Free
-
C-terminal Modification
- Free
-
Nonterminal Modifications and Unusual Amino Acids
- Free
-
Stereochemistry
- L
-
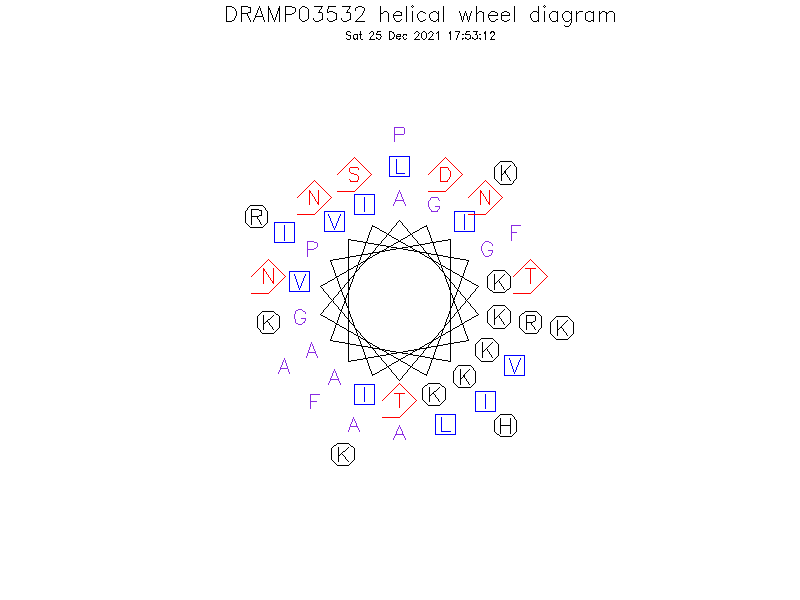
Structure
- Alpha helix (1 helices; 30 residues)
-
Structure Description
- The solution structure reveals an unique structure comprising of a long alpha-helix containing eight turns along nearly the full length of the peptide except for four N-terminal residues and six C-terminal residues. The electrostatic surface map shows that the N-terminal segment of the alpha-helix, residues 5-22, is an amphipathic alpha-helix with a clear separation of hydrophobic and hydrophilic faces, and that the C-terminal segment o
-
Helical Wheel Diagram
-
PDB ID
- 1KV4 resolved by NMR.
-
Predicted Structure
- There is no predicted structure for DRAMP03532.
Physicochemical Information
-
Formula
- C208H358N62O51
Absent Amino Acids
- CEMQWY
Common Amino Acids
- K
Mass
- 4543.52
PI
- 11.36
Basic Residues
- 12
Acidic Residues
- 1
Hydrophobic Residues
- 18
Net Charge
- +11
-
Boman Index
- -55.43
Hydrophobicity
- -0.21
Aliphatic Index
- 100
Half Life
-
- Mammalian:4.4 hour
- Yeast:>20 hour
- E.coli:>10 hour
Extinction Coefficient Cystines
- 0
Absorbance 280nm
- 0
Polar Residues
- 9
DRAMP03532
Comments Information
Function
- Has antibacterial activity against Gram-positive and Gram-negative bacteria. Probably acts by disturbing membrane functions with its amphipathic structure.
Literature Information
- ·Literature 1
-
Title
- Moricin, a novel type of antibacterial peptide isolated from the silkworm, Bombyx mori.
-
Pubmed ID
- 8530391
-
Reference
- J Biol Chem. 1995 Dec 15;270(50):29923-29927.
-
Author
- Hara S, Yamakawa M.
- ·Literature 2
-
Title
- Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori.
-
Pubmed ID
- 11997013
-
Reference
- FEBS Lett. 2002 May 8;518(1-3):33-38.
-
Author
- Hemmi H, Ishibashi J, Hara S, Yamakawa M.