General Information
-
DRAMP ID
- DRAMP04075
-
Peptide Name
- Antimicrobial peptide HP (2-20)
-
Source
- Synthetic construct (amino acid substitution)
-
Family
- Not found
-
Gene
- rplA
-
Sequence
- AKKVFKRLEKLFSKIQNDK
-
Sequence Length
- 19
-
UniProt Entry
- P56029
-
Protein Existence
- Synthetic
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram+, Anti-Gram-, Antifungal
-
Target Organism
-
- [Ref.16255716]Gram-positive bacteria: Bacillus subtilis (MIC=3.13 µg/ml), Staphylococcus epidermidis (MIC=6.25 µg/ml), Staphylococcus aureus (MIC=12.5 µg/ml), Listeria monocytogenes (MIC=6.25 µg/ml);
- Gram-negative bacteria: Escherichia coli (MIC=3.13 µg/ml), Salmonella typhimurium (MIC=1.56 µg/ml), Proteus vulgaris (MIC=6.25 µg/ml), Pseudomonas aeruginosa (MIC=6.25 µg/ml), Escherichia coli O157 (MIC=12.5 µg/ml);
- Fungi: Trichosporon beigelii (MIC=3.13 µg/ml), Candida albicans (MIC>25 µg/ml), Saccharomyces cerevisiae (MIC>25 µg/ml), Aspergillus flavus (MIC=50 µg/ml), Aspergillus fumigatus (MIC>50 µg/ml), Botrytis cinerea (MIC=12.5 µg/ml), Fusarium moniliforme (MIC=25 µg/ml).
-
Hemolytic Activity
-
- [Swiss_Prot Entry P56029]Non-hemolytic against mammalian cells
-
Cytotoxicity
-
- Not included yet
-
Binding Target
- Phospholipids
Structure Information
-
Linear/Cyclic
- Not included yet
-
N-terminal Modification
- Not included yet
-
C-terminal Modification
- Not included yet
-
Nonterminal Modifications and Unusual Amino Acids
- Not included yet
-
Stereochemistry
- Not included yet
-
Structure
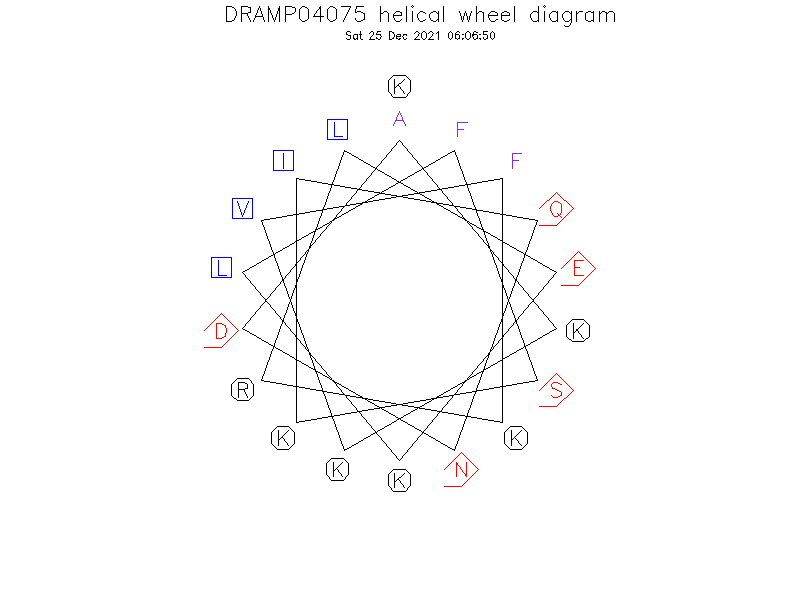
- Alpha helix (1 helices; 11 residues)
-
Structure Description
- Not found
-
Helical Wheel Diagram
-
PDB ID
- 1P0G resolved by NMR.
-
Predicted Structure
- There is no predicted structure for DRAMP04075.
Physicochemical Information
-
Formula
- C107H182N30O27
Absent Amino Acids
- CGHMPTWY
Common Amino Acids
- K
Mass
- 2320.81
PI
- 10.3
Basic Residues
- 7
Acidic Residues
- 2
Hydrophobic Residues
- 7
Net Charge
- +5
-
Boman Index
- -52.76
Hydrophobicity
- -1
Aliphatic Index
- 82.11
Half Life
-
- Mammalian:4.4 hour
- Yeast:>20 hour
- E.coli:>10 hour
Extinction Coefficient Cystines
- 0
Absorbance 280nm
- 0
Polar Residues
- 2
DRAMP04075
Comments Information
Function
- Peptides originating from the N-terminal end of L1 have antibacterial activity against bacteria such as E. coli and Bacillus megaterium and modest antifungal activities. Has no effect on H. pylori itself. Peptides are not hemolytic against mammalian cells. These peptides may be released in the stomach during altruistic lysis to kill other fast growing bacteria.
Literature Information
- ·Literature 1
-
Title
- Interactions between the plasma membrane and the antimicrobial peptide HP (2-20) and its analogues derived from Helicobacter pylori.
-
Pubmed ID
- 16255716
-
Reference
- Biochem J. 2006 Feb 15;394(Pt 1):105-114.
-
Author
- Lee KH, Lee DG, Park YK, Harm KS, Kim YM.
- ·Literature 2
-
Title
- Structure of antimicrobial peptide, HP (2-20) and its analogues derived from Helicobacter pylori, as determined by 1H NMR spectroscopy.
-
Pubmed ID
- PubMed ID is not available
-
Reference
- Submitted (SEP-2004) to the PDB data bank.
-
Author
- Lee KH, Lee DG, Park YK, Harm KS, Kim YM.
- ·Literature 3
-
Title
- Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy.
-
Pubmed ID
- PubMed ID is not available
-
Reference
- To be published
-
Author
- Lee KH, Lee DG, Park YK, Harm KS, Kim YM.