General Information
-
DRAMP ID
- DRAMP18533
-
Peptide Name
- V13KL (V681 peptide derivative)
-
Source
- Synthetic construct
-
Family
- Derived from the framework peptide V681
-
Gene
- Not found
-
Sequence
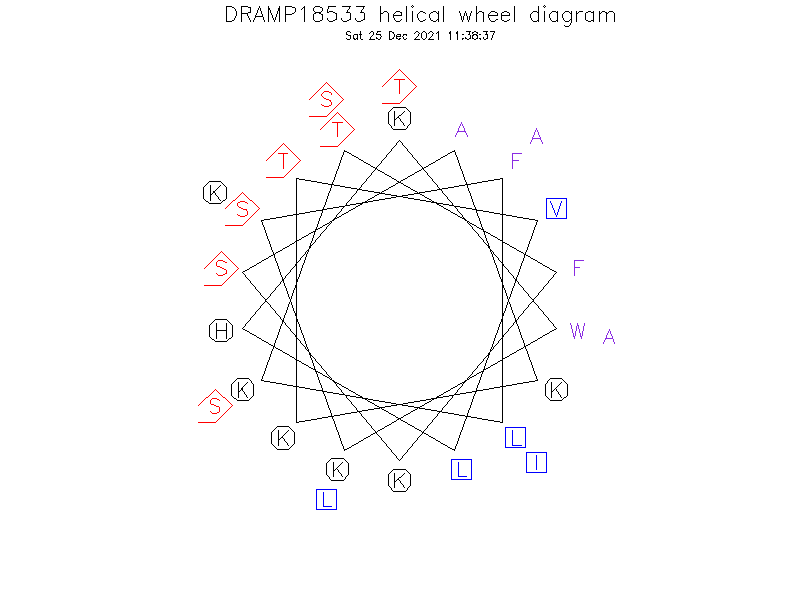
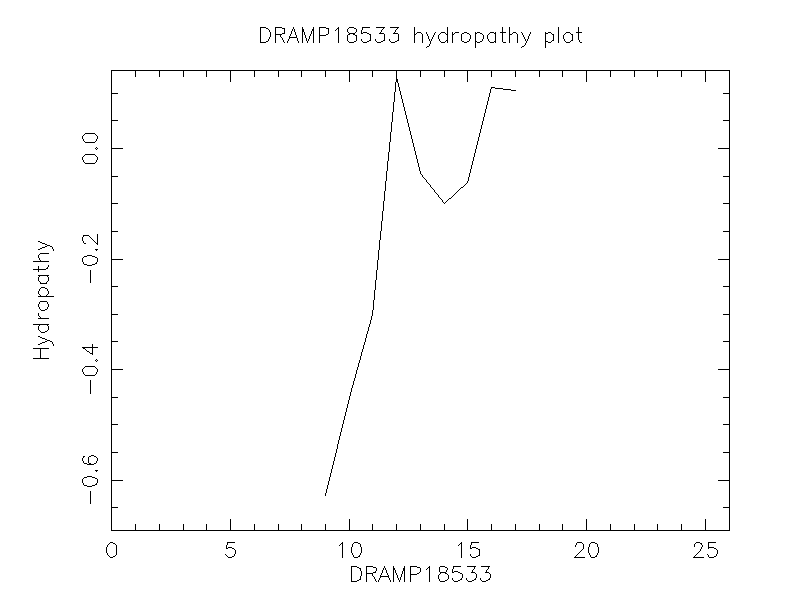
- KWKSFLKTFKSAKKTVLHTALKAISS
-
Sequence Length
- 26
-
UniProt Entry
- No entry found
-
Protein Existence
- Synthetic
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram+, Anti-Gram-
-
Target Organism
-
- [Ref.15677462]Gram-positive bacteria: Staphylococcus aureus 25923 (MIC=64.0 μg/ml), Staphylococcus aureus SAP0017 (MIC=64.0 μg/ml), Staphylococcus epidermidis C621 (MIC=5.0 μg/ml), Bacillus subtilis C971 (MIC=1.6 μg/ml), Enterococcus faecalis C625 (MIC=64.0 μg/ml), Corynebacterium xerosis C875 (MIC=1.3 μg/ml), The geometric mean of MIC values above six Gram-positive strains (MIC=11.8 μg/ml);
- Gram-negative bacteria: Escherichia coli UB1005 (MIC=2.5 μg/ml), Escherichia coli DC2 (MIC=1.6 μg/ml), Salmonella typhimurium C587 (MIC=4.0 μg/ml), Salmonella typhimurium C610 (MIC=1.3 μg/ml), Pseudomonas aeruginosa H187 (MIC=8.0 μg/ml), Pseudomonas aeruginosa H188 (MIC=5.0 μg/ml), The geometric mean of MIC values above six Gram-negative strains (MIC=3.1 μg/ml).
- [Ref.17158938]Gram-negative bacteria: Pseudomonas aeruginosa PAO1(MIC=31.3 μg/ml), Pseudomonas aeruginosa WR5 (MIC=250 μg/ml), Pseudomonas aeruginosa PAK (MIC=125 μg/ml), Pseudomonas aeruginosa PA14 (MIC=62.5 μg/ml), Pseudomonas aeruginosa M2 (MIC=31.3 μg/ml), Pseudomonas aeruginosa CP204 (MIC=7.8 μg/ml), The geometric mean of MIC values above six Pseudomonas aeruginosa strains (MIC=49.6 μg/ml)
-
Hemolytic Activity
-
- [Ref.17158938]MHC=250.0 μg/ml against human red blood cells
-
Cytotoxicity
-
- Not included yet
-
Binding Target
- Not found
Structure Information
-
Linear/Cyclic
- Not included yet
-
N-terminal Modification
- Not included yet
-
C-terminal Modification
- Not included yet
-
Nonterminal Modifications and Unusual Amino Acids
- Not included yet
-
Stereochemistry
- Not included yet
-
Structure
- Alpha helix
-
Structure Description
- Not found
-
Helical Wheel Diagram
-
PDB ID
- None
-
Predicted Structure
- There is no predicted structure for DRAMP18533.
Physicochemical Information
-
Formula
- C139H230N36O34
Absent Amino Acids
- CDEGMNPQRY
Common Amino Acids
- K
Mass
- 2949.58
PI
- 10.78
Basic Residues
- 8
Acidic Residues
- 0
Hydrophobic Residues
- 11
Net Charge
- +8
-
Boman Index
- 1.05
Hydrophobicity
- -0.215
Aliphatic Index
- 82.69
Half Life
-
- Mammalian:1.3 hour
- Yeast:3 min
- E.coli:2 min
Extinction Coefficient Cystines
- 5500
Absorbance 280nm
- 220
Polar Residues
- 7
DRAMP18533
Comments Information
Function
- Antibacterial activity against Gram-positive and Gram-negative bacteria. L-Lysine replace L-Valine at the position 13 on the non-polar face of peptide V681. Has hemolytic activity.
Literature Information
- ·Literature 1
-
Title
- Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index.
-
Pubmed ID
- 15677462
-
Reference
- J Biol Chem. 2005 Apr 1;280(13):12316-29.
-
Author
- Chen Y, Mant CT, Farmer SW, Hancock RE, Vasil ML, Hodges RS.
- ·Literature 2
-
Title
- Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides.
-
Pubmed ID
- 17158938
-
Reference
- Antimicrob Agents Chemother. 2007 Apr;51(4):1398-406.
-
Author
- Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS.