General Information
-
DRAMP ID
- DRAMP18666
-
Peptide Name
- P1 (Pilosulin-1 1-20; Ant, insects, arthropods, invertebrates, animals)
-
Source
- Myrmecia pilosula (Jumper ant)
-
Family
- Derived from the peptide Pilosulin-1, Belongs to the pilosulin family.
-
Gene
- Not found
-
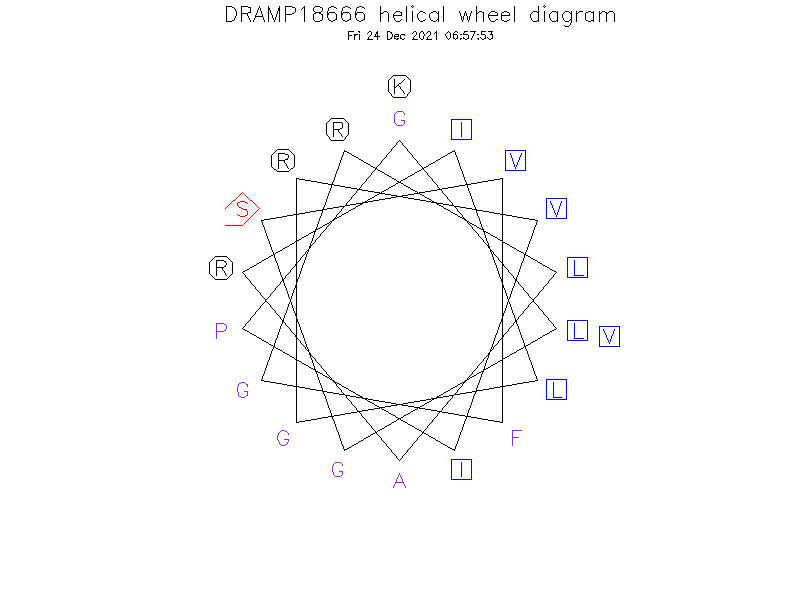
Sequence
- GLGSVFGRLARILGRVIPKV
-
Sequence Length
- 20
-
UniProt Entry
- Q07932
-
Protein Existence
- Protein level
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram+, Anitifungal, Anti-Gram-
-
Target Organism
-
- [Ref.15639237]Gram-positive bacteria: Staphylococcus aureus 710A (MIC=4 μM), Bacillus megaterium BM11 (MIC=2 μM);
- Gram-negative bacteria: Escherichia coli ML-35 (MIC=2 μM), Pseudomonas aeruginosa ATCC27853 (MIC=8 μM), Salmonella typhimurium ATCC14028 (MIC=8 μM).
- Drug resistant clinical isolates: Staphylococcus simulans 22 (MIC=2 μM), Micrococcus luteus (MIC=0.5 μM), Enterococcus spp. I-11305b (MIC=2 μM), Enterococcus spp. I-11054 (MIC=2 μM), Staphylococcus haemolyticus I-10925 (MIC=2 μM), Staphylococcus epidermidis LT1324 (MIC=2 μM), Staphylococcus aureus 5185 (MIC=2 μM), Staphylococcus aureus I-11574 (MIC=2 μM), Staphylococcus aureus LT1338 (MIC=3 μM), Staphylococcus aureus T1334 (MIC=1.5 μM), Citrobacter freundii I-11090 (MIC=2 μM), Klebsiella pneumoniae I-10910 (MIC=2 μM), Escherichia coli I-11276b (MIC=4 μM), Escherichia coli O-19592 (MIC=2 μM), Stenotrophomonas maltophilia O-16451 (MIC=2 μM), Stenotrophomonas maltophilia I-10717 (MIC=2 μM), Pseudomonas aeruginosa 4991 (MIC=4 μM), Pseudomonas aeruginosa I-10968 (MIC=4 μM), Candida albicans I-11301 (MIC=4 μM).
-
Hemolytic Activity
-
- [Ref.15639237]20% hemolysis at 10μM, 100% hemolysis at 100μM against human red blood cells
-
Cytotoxicity
-
- Not included yet
-
Binding Target
- Not found
Structure Information
-
Linear/Cyclic
- Not included yet
-
N-terminal Modification
- Not included yet
-
C-terminal Modification
- Not included yet
-
Nonterminal Modifications and Unusual Amino Acids
- Not included yet
-
Stereochemistry
- Not included yet
-
Structure
- Alpha helix
-
Structure Description
- Not found
-
Helical Wheel Diagram
-
PDB ID
- 5X3L resolved by NMR.
-
Predicted Structure
- There is no predicted structure for DRAMP18666.
Physicochemical Information
-
Formula
- C97H170N30O22
Absent Amino Acids
- CDEHMNQTWY
Common Amino Acids
- G
Mass
- 2108.6
PI
- 12.3
Basic Residues
- 4
Acidic Residues
- 0
Hydrophobic Residues
- 10
Net Charge
- +4
-
Boman Index
- -844
Hydrophobicity
- 0.81
Aliphatic Index
- 146
Half Life
-
- Mammalian:30 hour
- Yeast:>20 hour
- E.coli:>10 hour
Extinction Coefficient Cystines
- 0
Absorbance 280nm
- 0
Polar Residues
- 5
DRAMP18666
Comments Information
Function
- Has strong cytotoxic and hemolytic activities. Is more potent against mononuclear leukocytes than against granulocytes. Antimicrobial activity against Gram-positive bacteria, Gram-negative bacteria and fungi. Adopts an alpha-helical structure.
Allergenic properties
- Causes an allergic reaction in human. Binds to IgE.
Literature Information
- ·Literature 1
-
Title
- Identification and optimization of an antimicrobial peptide from the ant venom toxin pilosulin.
-
Pubmed ID
- 15639237
-
Reference
- Arch Biochem Biophys. 2005 Feb 15;434(2):358-64.
-
Author
- Zelezetsky I, Pag U, Antcheva N, Sahl HG, Tossi A.