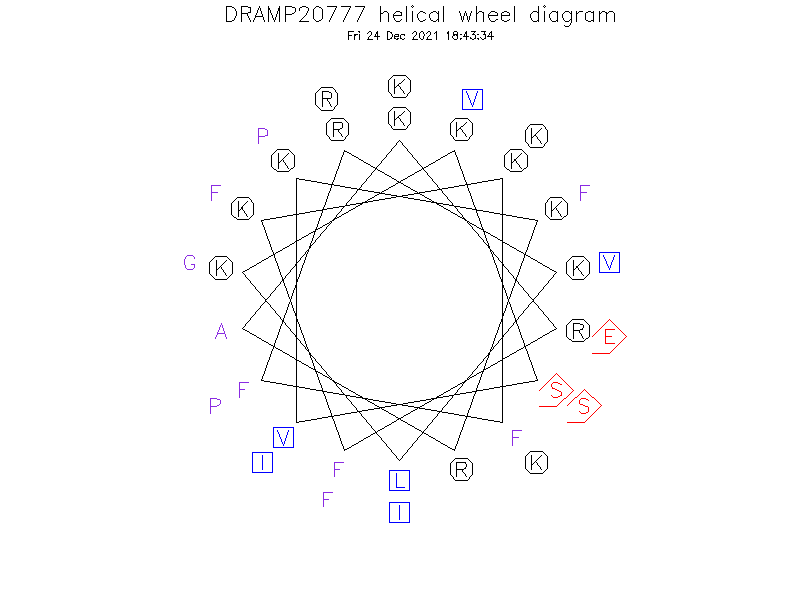General Information
-
DRAMP ID
- DRAMP20777
-
Peptide Name
- Cath-BF
-
Source
- Bungarus fasciatus (Banded krait)
-
Family
- Derived from three-finger toxin
-
Gene
- Not found
-
Sequence
- KRFKKFFRKLKKSVKKRAKEFFKKPRVIGVSIPF
-
Sequence Length
- 34
-
UniProt Entry
- No entry found
-
Protein Existence
- Protein level
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram+, Anti-Gram-
-
Target Organism
-
- [Ref.29392557] Gram-positive bacteria: MRSA PS-2 (MIC=64 μg/ml);
- Gram-negative bacteria, MRSA PS-7 (MIC=64 μg/ml), MRSA PS-18 (MIC=64 μg/ml), MRSA PS-45 (MIC=64 μg/ml), MRSA PS-46 (MIC=128 μg/ml), MRSA PS-50 (MIC=64 μg/ml), MRSA PS-59 (MIC=128 μg/ml), MRSA PS-70 (MIC=32 μg/ml), MRSA PS-84 (MIC=128 μg/ml), MRSA PS-106 (MIC=128 μg/ml), MRSA PS-131 (MIC=64 μg/ml), MRSA PS-139 (MIC=64 μg/ml), MRSA PS-162 (MIC=64 μg/ml), MRSA PS-165 (MIC=32 μg/ml), Staphylococcus aureus ATCC 25923 (MIC=16 μg/ml), Staphylococcus aureus ATCC 33591 (MIC=256 μg/ml);
- Gram-negative bacteria: Escherichia coli ATCC 25922 (MIC=8 μg/ml), Escherichia coli ATCC 51446 (MIC=16 μg/ml), Klebsiella pneumoniae ATCC 700603 (MIC=32 μg/ml), Klebsiella pneumoniae ATCC 13883 (MIC=4 μg/ml)
-
Hemolytic Activity
-
- [Ref.29392557] 5% haemolysis at 1.0 μg/ml, 5% haemolysis at 2.0 μg/ml, 5% haemolysis at 4.0 μg/ml, 5% haemolysis at 8.0 μg/ml, 7% haemolysis at 16.0 μg/ml, 9% haemolysis at 31.2 μg/ml, 13% haemolysis at 62.5 μg/ml, 17% haemolysis at 125.0 μg/ml, 19% haemolysis at 250.0 μg/ml, 23% haemolysis at 500.0 μg/ml, 24% haemolysis at 1000.0 μg/ml against human red blood cells
-
Cytotoxicity
-
- [Ref.29392557] The cell viability of HUVEC at the peptide concentration of 35.5 μg/ml, 71 μg/ml and 113 μg/ml is 127%, 100% and 76%.
- The cell viability of H9c2 at the peptide concentration of 35.5, 71, and 113 μg/ml is 132, 100, and 80%.
-
Binding Target
- Not found
Structure Information
-
Linear/Cyclic
- Linear
-
N-terminal Modification
- Free
-
C-terminal Modification
- Free
-
Nonterminal Modifications and Unusual Amino Acids
- None
-
Stereochemistry
- L
-
Structure
- Alpha helix
-
Structure Description
- Not found
-
Helical Wheel Diagram
-
PDB ID
- None
-
Predicted Structure
- There is no predicted structure for DRAMP20777.
Physicochemical Information
-
Formula
- C203H335N57O39
Absent Amino Acids
- CDHMNQTWY
Common Amino Acids
- K
Mass
- 4198.25
PI
- 12.04
Basic Residues
- 15
Acidic Residues
- 1
Hydrophobic Residues
- 13
Net Charge
- +14
-
Boman Index
- -8683
Hydrophobicity
- -0.753
Aliphatic Index
- 62.94
Half Life
-
- Mammalian:1.3 hour
- Yeast:3 min
- E.coli:2 min
Extinction Coefficient Cystines
- 0
Absorbance 280nm
- 0
Polar Residues
- 3
DRAMP20777
Comments Information
Function
- Antibacterial activity against the Gram-positive and Gram-negative bacteria.
Literature Information
- ·Literature 1
-
Title
- The antimicrobial potential of a new derivative of cathelicidin from Bungarus fasciatus against methicillin-resistant Staphylococcus aureus.
-
Pubmed ID
- 29392557
-
Reference
- J Microbiol. 2018 Feb;56(2):128-137.
-
Author
- Tajbakhsh M, Karimi A, Tohidpour A, Abbasi N, Fallah F, Akhavan MM.