General Information
-
DRAMP ID
- DRAMP20838
-
Peptide Name
- Pb-CATH4 bivittatus antimicrobial peptides peptide derivative
-
Source
- Synthetic construct
-
Family
- Derived from cathelicidin family
-
Gene
- Not found
-
Sequence
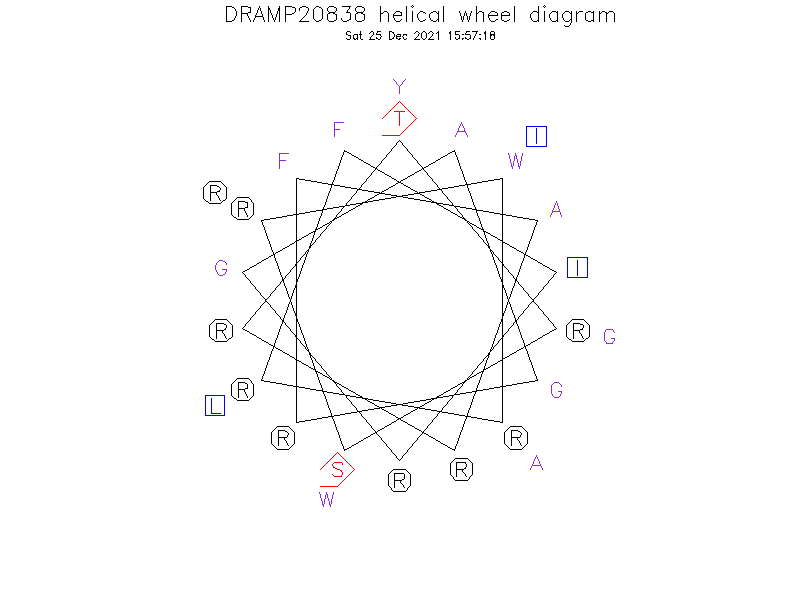
- TRSRWRRFIRGAGRFARRYGWRIAL
-
Sequence Length
- 25
-
UniProt Entry
- No entry found
-
Protein Existence
- Synthetic
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram+, Anti-Gram-
-
Target Organism
-
- [Ref.28630199]Gram-negative bacteria:Escherichia coli (ATCC 25922)(MIC=1μg/ml);Pseudomonas aeruginosa (ATCC 27853)(MIC=3μg/ml);S. typhimurium (ATCC 14028)(MIC=0.5μg/ml);
- Gram-positive bacteria:Staphylococcus aureus (ATCC 29213)(MIC>128μg/ml);B. cereus (ATCC 10876)(MIC=10μg/ml);
- Clinical isolates:Escherichia coli(MIC=4.7μg/ml);K. pneumoniae(MIC=1.18μg/ml)
-
Hemolytic Activity
-
- [Ref.28630199] 0% hemolysis at 4μg/ml against human red blood cell; 1.4 ± 0.4% hemolysis at 8μg/ml against human red blood cell; 5.1 ± 0.5% hemolysis at 16μg/ml against human red blood cell;7.3 ± 0.6% hemolysis at 32μg/ml against human red blood cell; 12.2 ± 0.7% hemolysis at 64μg/ml against human red blood cell
-
Cytotoxicity
-
- [Ref.28630199] The cell viability of Pk15(pig kidney fibroblasts) is 103.8%, 124.6%, 94.7%,55.2% and 53.9% at peptide concentrations of 5, 10, 20, 30 and 64 μg/ml.
- The cell viability of HaCaT(human keratinocytes) is 106.9%, 94.6%, 66.2%, 58.1% and 51.
-
Binding Target
- Not found
Structure Information
-
Linear/Cyclic
- Linear
-
N-terminal Modification
- Free
-
C-terminal Modification
- Free
-
Nonterminal Modifications and Unusual Amino Acids
- None
-
Stereochemistry
- L
-
Structure
- Alpha helix
-
Structure Description
- The peptide adopts arginine content of this peptide is 37.5%, and it contains two tryptophan residues (8.3%)
-
Helical Wheel Diagram
-
PDB ID
- None
-
Predicted Structure
- There is no predicted structure for DRAMP20838.
Physicochemical Information
-
Formula
- C143H226N54O29
Absent Amino Acids
- CDEHKMNPQV
Common Amino Acids
- R
Mass
- 3165.71
PI
- 12.54
Basic Residues
- 9
Acidic Residues
- 0
Hydrophobic Residues
- 10
Net Charge
- +9
-
Boman Index
- -10676
Hydrophobicity
- -0.9
Aliphatic Index
- 58.8
Half Life
-
- Mammalian:7.2 hour
- Yeast:>20 hour
- E.coli:>10 hour
Extinction Coefficient Cystines
- 12490
Absorbance 280nm
- 520.42
Polar Residues
- 6
DRAMP20838
Comments Information
Function
- Antibacterial activity against the Gram-positive and Gram-negative bacteria
Literature Information
- ·Literature 1
-
Title
- Genomewide Analysis of the Antimicrobial Peptides in Python bivittatus and Characterization of Cathelicidins with Potent Antimicrobial Activity and Low Cytotoxicity.
-
Pubmed ID
- 28630199
-
Reference
- Antimicrob Agents Chemother. 2017 Aug 24;61(9).
-
Author
- Kim D, Soundrarajan N, Lee J, Cho HS, Choi M, Cha SY, Ahn B, Jeon H, Le MT, Song H, Kim JH, Park C.