General Information
-
DRAMP ID
- DRAMP21088
-
Peptide Name
- PR-TR (Derived from PRW4)
-
Source
- Synthetic construct
-
Family
- Not found
-
Gene
- Not found
-
Sequence
- RFRRLRWKTRWRLKKIWWPFLRR
-
Sequence Length
- 23
-
UniProt Entry
- No entry found
-
Protein Existence
- Synthetic form
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram+, Anti-Gram-
-
Target Organism
-
- [Ref.25818457] Gram-positive bacteria : Staphylococcus aureus 25923(MIC=26.7 mg/L), Staphylococcus aureus 29213(MIC=6.7 mg/L), Staphylococcus epidermidis 12228(MIC=13.4 mg/L), Enterococcus faecalis 29212(MIC=3.3 mg/L), Bacillus subtilis 11060(MIC=3.3 mg/L);
- Gram-negative bacteria : Escherichia coli 078(MIC=13.4 mg/L), Escherichia coli K88(MIC=1.7 mg/L), Escherichia coli ER2566(MIC=3.3 mg/L), Escherichia coli 44102(MIC=6.7 mg/L), Escherichia coli 1515(MIC=1.7 mg/L), Escherichia coli 25922(MIC=6.7 mg/L), Escherichia coli UB1005(MIC=3.3 mg/L), Salmonella typhimurium 7731(MIC=6.7 mg/L), Salmonella typhimurium 1402(MIC=13.4 mg/L), Pseudomonas aeruginosa 21625(MIC=6.7 mg/L), Pseudomonas aeruginosa 21630(MIC=13.4 mg/L), Pseudomonas aeruginosa 10419(MIC=6.7 mg/L), Pseudomonas aeruginosa 27853(MIC=13.4 mg/L), Salmonella pullorum 7913(MIC=13.4 mg/L)
-
Hemolytic Activity
-
- [Ref.25818457] 0% hemolysis at 9.2 mg/L , 1% hemolysis at 18.4 mg/L , 4% hemolysis at 36.8mg/L , 10% hemolysis at 73.6 mg/L , 17% hemolysis at 147.1 mg/L , 22% hemolysis at 294.3 mg/L against human red blood cells
-
Cytotoxicity
-
- [Ref.25818457] The cell viability of PR-TR is 94%, 89%, 83%, 77%, 74%, 71%, 68%, 66% and 60% at peptide concentrations of 1.1, 2.3, 4.6, 9.2, 18.4, 36.8, 73.6, 147.1 and 294.3 μg/ml
-
Binding Target
- Not found
Structure Information
-
Linear/Cyclic
- Linear
-
N-terminal Modification
- Free
-
C-terminal Modification
- Amidation
-
Nonterminal Modifications and Unusual Amino Acids
- None
-
Stereochemistry
- L
-
Structure
- Random coil, alpha helix, beta Sheet
-
Structure Description
- Not found
-
Helical Wheel Diagram
-
PDB ID
- None
-
Predicted Structure
- There is no predicted structure for DRAMP21088.
Physicochemical Information
-
Formula
- C161H250N54O25
Absent Amino Acids
- ACDEGHMNQSVY
Common Amino Acids
- R
Mass
- 3342.1
PI
- 12.85
Basic Residues
- 11
Acidic Residues
- 0
Hydrophobic Residues
- 10
Net Charge
- +11
-
Boman Index
- -10362
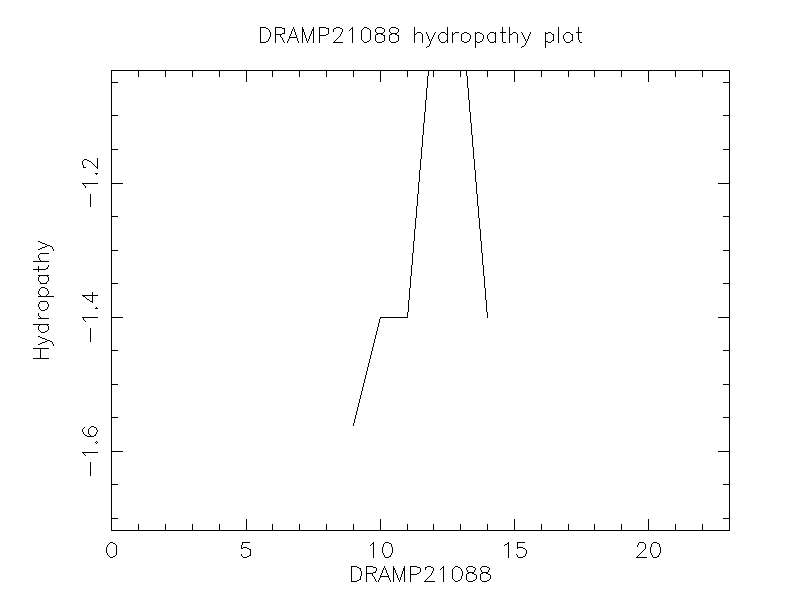
Hydrophobicity
- -1.396
Aliphatic Index
- 67.83
Half Life
-
- Mammalian:1 hour
- Yeast:2 min
- E.coli:2 min
Extinction Coefficient Cystines
- 22000
Absorbance 280nm
- 1000
Polar Residues
- 1
DRAMP21088
Comments Information
Function
- Antibacterial activity against Gram-positive bacteria and Gram-negative bacteria
Literature Information
- ·Literature 1
-
Title
- Characterization of cell selectivity, physiological stability and endotoxin neutralization capabilities of a-helix-based peptide amphiphiles
-
Pubmed ID
- 25818457
-
Reference
- Biomaterials. 2015 Jun;52:517-30. doi: 10.1016/j.biomaterials.2015.02.063.
-
Author
- Zhi Ma, Dandan Wei, Ping Yan, Xin Zhu, Anshan Shan , Zhongpeng Bi