General Information
-
DRAMP ID
- DRAMP21350
-
Peptide Name
- 2IH4 (De Novo Synthesis)
-
Source
- synthetic construct
-
Family
- Not found
-
Gene
- Not found
-
Sequence
- IHHIHHHIHHIHHHIHHIHHHIHHIHHH
-
Sequence Length
- 28
-
UniProt Entry
- No entry found
-
Protein Existence
- Synthetic form
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram-
-
Target Organism
-
- [Ref.31199117] Gram-negative Bacteria: Escherichia coli 25922 (MIC=1μM), Escherichia coli UB1005 (MIC=2μM), Escherichia coli K88 (MIC=4>64μM), Escherichia coli K99 (MIC=1μM), Escherichia coli 078 (MIC=4>64μM), Escherichia coli 987P (MIC=1μM), Pseudomonas aeruginosa 27853 (MIC=1μM), Pseudomonas aeruginosa PAO1 (MIC=1μM), Salmonella typhimurium 14028 (MIC=64μM), Salmonella typhimurium 7731 (MIC>64μM)
-
Hemolytic Activity
-
- [Ref.31199117] 5% hemolysis at 63μM, 8% hemolysis at 128μM against human red blood cells
-
Cytotoxicity
-
- [Ref.31199117] ①The cell viability of HEK293T cells induced by 2IH4 is at peptide concentrations of 2, 4, 8, 16, 32, 64 and 128 μM at pH 7.4, while that induced by Melittin is 47.2%, 1.7%, 1.9%, 0.6%, 0.6%, 0.6% and 1.7% at 2, 4, 8, 16, 32, 64 and 128 μM. ②The cell viability of HEK293T cells induced by 2IH4 is 103.7%, 103.7%, 104.8%, 103.4%, 105.3%, 94.9% and 57.9% at peptide concentrations of 2, 4, 8, 16, 32, 64 and 128 μM at pH 6.0, while that induced by Melittin is 43.8%, 3.4%, 3.4%, 0%, 2.2%, 0% and 1.1% at 2, 4, 8, 16, 32, 64 and 128 μM.
-
Binding Target
- Not found
Structure Information
-
Linear/Cyclic
- Linear
-
N-terminal Modification
- Free
-
C-terminal Modification
- Amidation
-
Nonterminal Modifications and Unusual Amino Acids
- None
-
Stereochemistry
- L
-
Structure
- 95% α-helix in 50% TFE/H2O
-
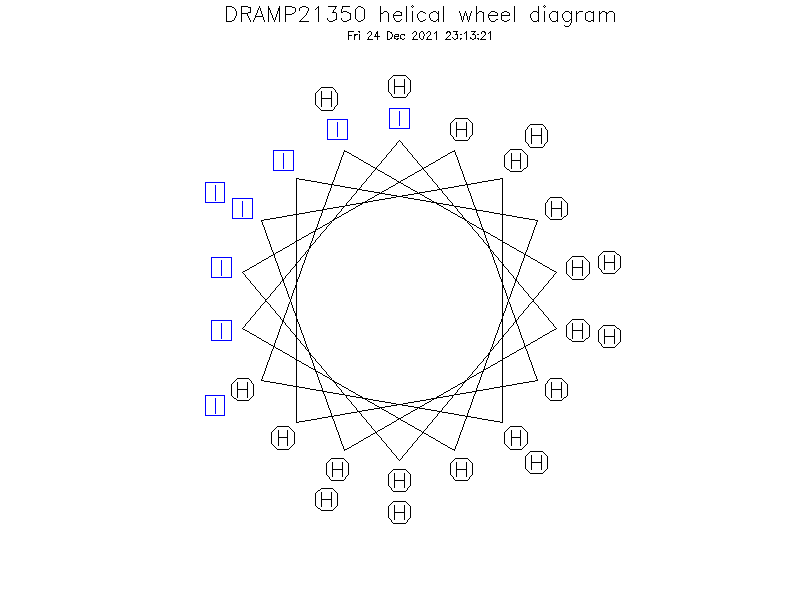
Structure Description
- The α-helical coiled coils would show a sequence periodicity of seven residues (heptad repeat), indicated as “abcdefg”, with hydrophobic residues in the “a” and “d” positions “forming the knobs-into-holes packing interactions and providing the energy needed to distort the mechanical-stabilized α-helices”.
-
Helical Wheel Diagram
-
PDB ID
- None
-
Predicted Structure
- There is no predicted structure for DRAMP21350.
Physicochemical Information
-
Formula
- C168H230N68O29
Absent Amino Acids
- ACDEFGKLMNPQRSTVW
Common Amino Acids
- H
Mass
- 3666.11
PI
- 7.55
Basic Residues
- 20
Acidic Residues
- 0
Hydrophobic Residues
- 8
Net Charge
- +20
-
Boman Index
- -5384
Hydrophobicity
- -1
Aliphatic Index
- 111.43
Half Life
-
- Mammalian:20 hour
- Yeast:30 min
- E.coli:>10 hour
Extinction Coefficient Cystines
- 0
Absorbance 280nm
- 0
Polar Residues
- 0
DRAMP21350
Comments Information
Function
- Antibacterial activity against Gram-negative bacteria.
Literature Information
- ·Literature 1
-
Title
- Highly Stabilized α-Helical Coiled Coils Kill Gram-Negative Bacteria by Multicomplementary Mechanisms under Acidic Condition
-
Pubmed ID
- 31199117
-
Reference
- ACS Appl Mater Interfaces. 2019 Jun 26;11(25):22113-22128. doi: 10.1021/acsami.9b04654. Epub 2019 Jun 14.
-
Author
- Lai Z, Tan P, Zhu Y, Shao C, Shan A, Li L.