General Information
-
DRAMP ID
- DRAMP21404
-
Peptide Name
- peptide 2 (De Novo Synthesis)
-
Source
- synthetic construct
-
Family
- Not found
-
Gene
- Not found
-
Sequence
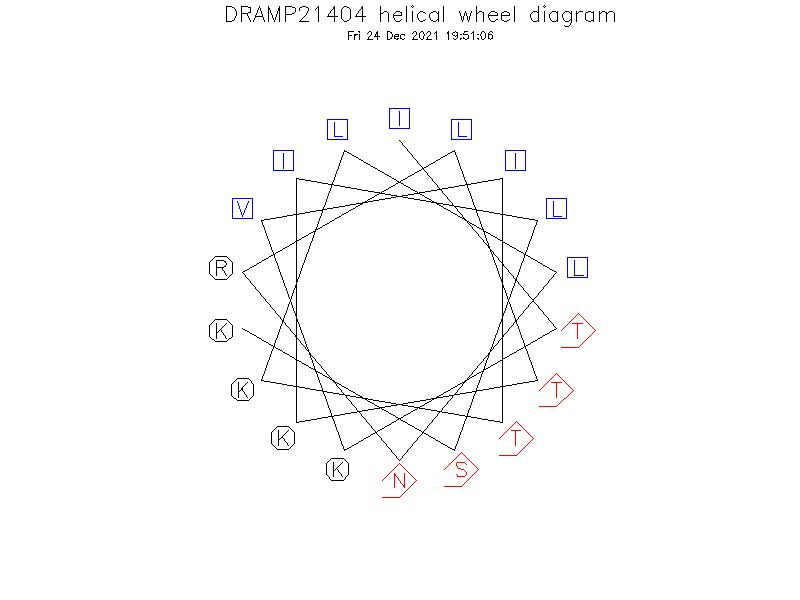
- ITKVITKLLNRLTKILSK
-
Sequence Length
- 18
-
UniProt Entry
- No entry found
-
Protein Existence
- Synthetic form
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram+, Anti-Gram-
-
Target Organism
-
- [Ref.31117348] Gram-positive bacteria:Staphylococcus aureus ATCC 6538(MIC=15 ± 2.5μM), Bacillus subtilis ATCC 6633(MIC=15 ± 2.5μM);
- Gram-negative bacteria:Escherichia coli ATCC 15597(MIC=3.1μM), Pseudomonas aeruginosa (MIC=25μM), Salmonella Typhimurium 6192(MIC=40 ± 5μM), Klebsiella pneumoniae NCTC 5055(MIC=50μM)
-
Hemolytic Activity
-
- [Ref.31117348] 5% hemolysis at 250 μM against human blood cells.
-
Cytotoxicity
-
- [Ref.31117348] ①The cell viability of MCF7 cells induced by peptide 2 is 94.2%, 88.6%, 82.0%, 55.0%, 44.0%, 32.4%, 15.3%, 15.8%, 12.8% and 1.2% at peptide concentrations of 1, 10, 15, 20, 25, 30, 35, 40, 50 and 100 μM. EC50=22.5±0.7 μM. ②The cell viability of Hela cells induced by peptide 2 is 96.1%, 95.4%, 83.8%, 64.2%, 57.7%, 49.5%, 31.5%, 30.2%, 7.8% and 0.2% at peptide concentrations of 1, 10, 15, 20, 25, 30, 35, 40, 50 and 100 μM. EC50=29.8±1 μM. ③The cell viabiity of HDF cells induced by peptide 2 is 96.3%, 89.7%, 89.9%, 92.7%, 95.0%, 93.4%, 93.1%, 96.1%, 88.6% and 67.4% at peptide concentrations of 1, 10, 15, 20, 25, 30, 35, 40, 50 and 100 μM.
-
Binding Target
- Not found
Structure Information
-
Linear/Cyclic
- Linear
-
N-terminal Modification
- Free
-
C-terminal Modification
- Free
-
Nonterminal Modifications and Unusual Amino Acids
- None
-
Stereochemistry
- L
-
Structure
- α-helix mainly in 10 mM phosphate buffer, pH7.4
-
Structure Description
- Not found
-
Helical Wheel Diagram
-
PDB ID
- None
-
Predicted Structure
- There is no predicted structure for DRAMP21404.
Physicochemical Information
-
Formula
- C96H180N26O24
Absent Amino Acids
- ACDEFGHMPQWY
Common Amino Acids
- KL
Mass
- 2082.64
PI
- 11.33
Basic Residues
- 5
Acidic Residues
- 0
Hydrophobic Residues
- 8
Net Charge
- +5
-
Boman Index
- -1639
Hydrophobicity
- 0.356
Aliphatic Index
- 167.78
Half Life
-
- Mammalian:20 hour
- Yeast:30 min
- E.coli:>10 hour
Extinction Coefficient Cystines
- 0
Absorbance 280nm
- 0
Polar Residues
- 5
DRAMP21404
Comments Information
Function
- Antibacterial activity against Gram-positive bacteria and Gram-nagetive bacteria.
Literature Information
- ·Literature 1
-
Title
- Helminth Defense Molecules as Design Templates for Membrane Active Antibiotics.
-
Pubmed ID
- 31117348
-
Reference
- ACS Infect Dis. 2019 May 30. doi: 10.1021/acsinfecdis.9b00157.
-
Author
- Hammond K, Lewis H, Faruqui N, Russell C, Hoogenboom BW, Ryadnov MG