General Information
-
DRAMP ID
- DRAMP29065
-
Peptide Name
- AMPR-11(derived from Romo1)
-
Source
- Homo sapiens (Human)
-
Family
- Belongs to the MGR2 family.
-
Gene
- ROMO1
-
Sequence
- KTMMQSGGTFGTFMAIGMGIR
-
Sequence Length
- 21
-
UniProt Entry
- P60602
-
Protein Existence
- Protein level
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram-, Anti-Gram+
-
Target Organism
-
- [Ref.33291307] Gram-positive bacteria: S. aureus(MBC=100 μg/mL), B. subtilis(MBC=90 μg/mL), E. faecium(MBC=85 μg/mL), S. sindenensis(MBC=95 μg/mL), E. faecalis(MBC=85 μg/mL), S. pneumoniae(MBC=100 μg/mL)
- Gram-negative bacteria: E. coli(MBC=85 μg/mL), P. aeruginosa(MBC=100 μg/mL), K. pneumoniae(MBC=100 μg/mL), A. baumannii(MBC=100 μg/mL), E. aerogenes(MBC=90 μg/mL)
- Multi-drug resistant bacteria: Methicillin-resistant S. aureus(MBC=100 μg/mL), Carbapenem-resistant P. aeruginosa(MBC=110 μg/mL), Carbapenem-resistant A. baumannii(MBC=110 μg/mL), Carbapenem-resistant K. pneumoniae(MBC=100 μg/mL), Vancomycin-resistant S. aureus(MBC=120 μg/mL), Vancomycin-resistant E. faecium(MBC=100 μg/mL )
-
Hemolytic Activity
-
- [Ref.33291307]At the concentration of 32 μg/mL, AMPR-11 induced a hemolysis of 20% on mouse red blood cells; At the concentration of 256 μg/mL, AMPR-11 induced a hemolysis of 23%.
-
Cytotoxicity
-
- [Ref.33291307] ①The cell viability of HeLa cells was 90%, 87%, 83% and 79% at the concentration of 32, 64, 128 and 256 μg/mL. ②The cell viability of HEK293 cells was 86%, 83%, 80% and 78% at the concentration of 32, 64, 128 and 256 μg/mL. ③The cell viability of HeLa cells was 89%, 86%, 83% and 80% at the concentration of 32, 64, 128 and 256 μg/mL.
-
Binding Target
- Bacterial membrane
Structure Information
-
Linear/Cyclic
- Linear
-
N-terminal Modification
- Free
-
C-terminal Modification
- Free
-
Nonterminal Modifications and Unusual Amino Acids
- None
-
Stereochemistry
- L
-
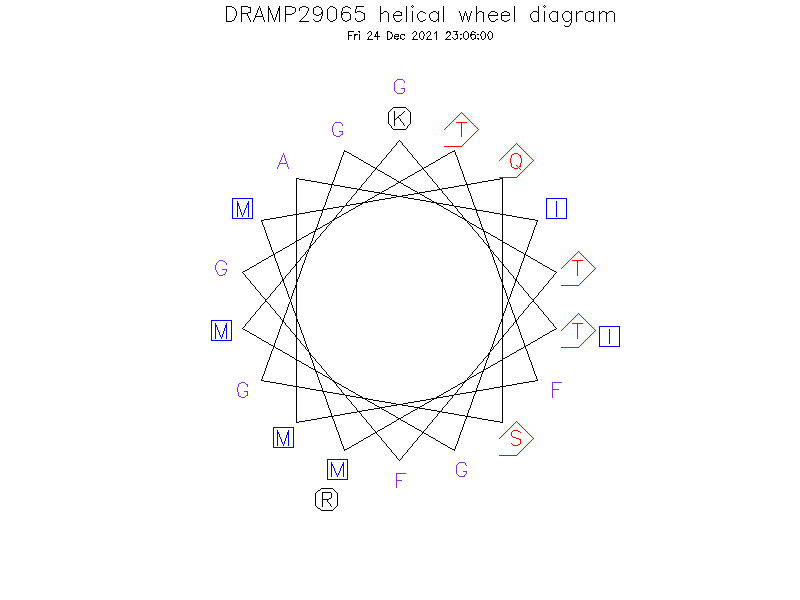
Structure
- α-helical
-
Structure Description
- AMPR-11 formed a random coil structure in distilled water but formed an ordered conformation in 50% hexafluoro-2-propanol (HFIP), which has been used for implementing a membrane-mimic environment
-
Helical Wheel Diagram
-
PDB ID
- None
-
Predicted Structure
- There is no predicted structure for DRAMP29065.
Physicochemical Information
-
Formula
- C95H156N26O27S4
Absent Amino Acids
- CDEHLNPVWY
Common Amino Acids
- G
Mass
- 2222.68
PI
- 11
Basic Residues
- 2
Acidic Residues
- 0
Hydrophobic Residues
- 5
Net Charge
- +2
-
Boman Index
- -541
Hydrophobicity
- 0.343
Aliphatic Index
- 41.9
Half Life
-
- Mammalian:1.3 hour
- Yeast:3 min
- E.coli:2 min
Extinction Coefficient Cystines
- 0
Absorbance 280nm
- 0
Polar Residues
- 9
DRAMP29065
Comments Information
Comment
- AMP derived from Romo1 (AMPR-11) is a promising agent for treatment of sepsis caused by MDR bacteria, and AMPR-11 could be combined with other potential antibiotics for clinical treatment.
Literature Information
- ·Literature 1
-
Title
- Romo1-Derived Antimicrobial Peptide Is a New Antimicrobial Agent against Multidrug-Resistant Bacteria in a Murine Model of Sepsis
-
Pubmed ID
- 32291307
-
Reference
- mBio. 2020 Apr 14;11(2):e03258-19. doi: 10.1128/mBio.03258-19.
-
Author
- Lee HR, You DG, Kim HK, Sohn JW, Kim MJ, Park JK, Lee GY, Yoo YD