General Information
-
DRAMP ID
- DRAMP29074
-
Peptide Name
- dCATH (duck cathelicidin)
-
Source
- Anas platyrhynchos platyrhynchos (Northern mallard)
-
Family
- Not found
-
Gene
- N/A
-
Sequence
- KRFWQLVPLAIKIYRAWKRR
-
Sequence Length
- 20
-
UniProt Entry
- A0A493U1S6
-
Protein Existence
- Synthetic form
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial, Anti-Gram-, Anti-Gram+
-
Target Organism
-
- [Ref.32698035] Gram-positive bacteria: S. aureus KCTC 1621 (MIC = 8 μM), S. epidermidis KCTC 1917 (MIC = 8 μM), B. subtilis KCTC 3068 (MIC = 8 μM);
- Resistant Gram-positive bacteria: MRSA CCARM 3089 (MIC = 16 μM), MRSA CCARM 3090 (MIC = 16 μM), MRSA CCARM 3095 (MIC = 8 μM), VREF ATCC 51559(MIC = 16 μM);
- Gram-negative bacteria: E. coli KCTC 1682 (MIC = 8 μM), P. aeruginosa KCTC 1637(MIC = 16 μM), S. typhimurium KCTC 1926 (MIC = 4 μM);
- Resistant Gram-negative bacteria: MDRPAd CCARM 2095 (MIC = 16 μM), MDRPA CCARM 2109 (MIC = 16 μM);
- Yeast: C. albicans KCTC 7965 (MIC = 8 μM), C. albicans KCTC 7121 (MIC = 16 μM).
-
Hemolytic Activity
-
- [Ref.32698035] HC10 = 5.1 μM. Note: HC10 is the peptide concentration that caused 10% hemolysis of sheep red blood cells (sRBCs).
-
Cytotoxicity
-
- [Ref.32698035] At the concentration of 1.25, 2.5, 5, 10, 20, 40 and 80 μM, the cell survival of dCATH against human bone marrow SH-SY5Y cells is 103%, 103%, 100%, 68%, 36%, 13% and 8%.
-
Binding Target
- Not found
Structure Information
-
Linear/Cyclic
- Linear
-
N-terminal Modification
- Free
-
C-terminal Modification
- Free
-
Nonterminal Modifications and Unusual Amino Acids
- None
-
Stereochemistry
- L
-
Structure
- Unknown
-
Structure Description
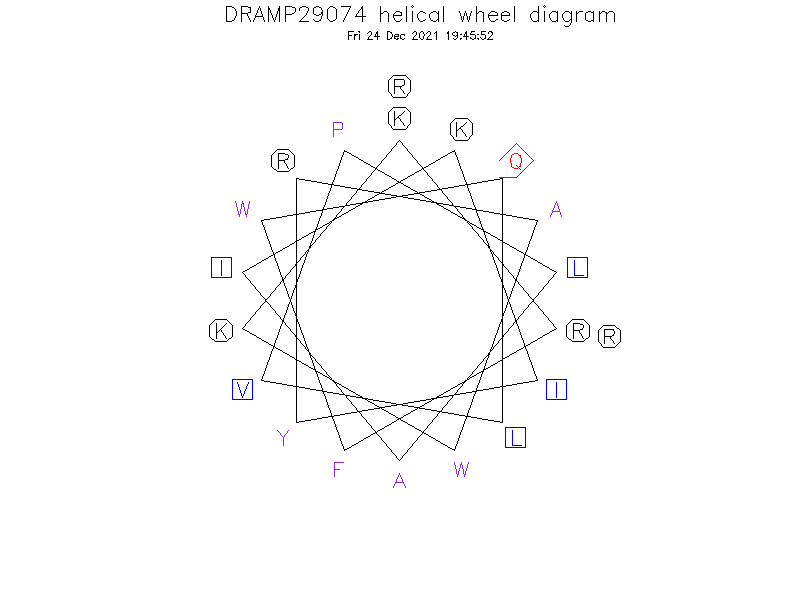
- Initially, four analog peptides were designed by truncating two, four, six, and eight amino acids at the N-terminal ends of dCATH peptide. dCATH and its truncated analogs could not adopt the amphipathic α-helical structure since the hydrophobic and charged residues were dispersed all around the molecule.
-
Helical Wheel Diagram
-
PDB ID
- None
-
Predicted Structure
- There is no predicted structure for DRAMP29074.
Physicochemical Information
-
Formula
- C127H202N38O23
Absent Amino Acids
- CDEGHMNST
Common Amino Acids
- R
Mass
- 2629.24
PI
- 12.02
Basic Residues
- 7
Acidic Residues
- 0
Hydrophobic Residues
- 10
Net Charge
- +7
-
Boman Index
- -4703
Hydrophobicity
- -0.535
Aliphatic Index
- 102.5
Half Life
-
- Mammalian:1.3 hour
- Yeast:3 min
- E.coli:2 min
Extinction Coefficient Cystines
- 12490
Absorbance 280nm
- 657.37
Polar Residues
- 1
DRAMP29074
Comments Information
Comment
- The peptide induces potent antimicrobial activity.
Literature Information
- ·Literature 1
-
Title
- Antimicrobial and anti-inflammatory activities of short dodecapeptides derived from duck cathelicidin: Plausible mechanism of bactericidal action and endotoxin neutralization
-
Pubmed ID
- 32698035
-
Reference
- Eur J Med Chem. 2020 Oct 15;204:112580. doi: 10.1016/j.ejmech.2020.112580. Epub 2020 Jul 16.
-
Author
- Kumar SD, Shin SY