General Information
-
DRAMP ID
- DRAMP29099
-
Peptide Name
- CAP18
-
Source
- Oryctolagus cuniculus
-
Family
- Not found
-
Gene
- camp
-
Sequence
- GLRKRLRKFRNKIKEKLKKIGQKIQGLLPKLAPRTDY
-
Sequence Length
- 37
-
UniProt Entry
- G1T9L4
-
Protein Existence
- Homology
Activity Information
-
Biological Activity
- Antimicrobial, Antibacterial,Anti-Gram+,Anti-Gram-
-
Target Organism
-
- [Ref.11557478]Gram-negative bacteria:Pseudomonas aeruginosa(MIC=1-32 µg/ml); Pseudomonas aeruginosa(MIC50=4 µg/ml); Pseudomonas aeruginosa(MIC90=16 µg/ml); Stenotrophomonas maltophilia(MIC=2-16 µg/ml); Achromobacter xylosoxidans(MIC=2-32 µg/ml).
- [Ref.10768969]Gram-positive bacteria:Staphylococcus aureus ATCC 33591(MIC=1.24 µM ); Staphylococcus aureus ATCC 33591(100 mM NaCl,MIC=1.36 µM); Staphylococcus aureus 930918-3(MIC=0.41 µM); Staphylococcus aureus 930918-3(100 mM NaCl,MIC=0.63 µM);
- Gram-negative bacteria:Pseudomonas aeruginosa PAO1(25 mM NaCl,EC50=0.22±0.05 µM); Pseudomonas aeruginosa PAO1(175 mM NaCl, EC50=0.11±0.02 µM); Escherichia coli DH5α(MIC=0.19 µM); Escherichia coli DH5α(100 mM NaCl, MIC=0.21 µM); Escherichia coli ML-35p(MIC=0.05 µM); Escherichia coli ML-35p(100 mM NaCl, MIC=0.09 µM); Pseudomonas aeruginosa PAO1(MIC=0.20 µM); Pseudomonas aeruginosa PAO1(100 mM NaCl, MIC=0.62 µM); Pseudomonas aeruginosa MR3007(MIC=0.05 µM); Pseudomonas aeruginosa MR3007(100 mM NaCl, MIC=0.36 µM).
- [Ref.26656394]Gram-negative bacteria:Yersinia ruckeri 392/2003(MIC=2-4 µg/ml); Aeromonas salmonicida ATCC 49385(MIC=2 µg/ml); Salmonella enterica serovar Typhimurium LT2(MIC=4-8 µg/ml); Campylobacter jejuni NCTC 11168(MIC=1 µg/ml); Escherichia coli ATCC 25922(MIC=4-8 µg/ml ); Pseudomonas aeruginosa ATCC 27853(MIC=4-8 µg/ml); Escherichia coli BW25113(MIC=4-8 µg/ml); Escherichia coli BW25113 del-rfaC(MIC=4 µg/ml); Escherichia coli ATCC 25922 del-rfaC(MIC=2 µg/ml);
- Gram-positive bacterial:Flavobacterium psychrophilum 1947(MIC>>256 µg/ml); Staphylococcus aureus ATCC 29213(MIC>=32 µg/ml); Enterococcus faecalis ATCC 29212(MIC=8 µg/ml); Listeria monocytogenes N22-2(MIC=2-4 µg/ml).
- [Ref.30245684]Gram-negative bacterial:Escherichia coli ATCC 25922 del-waaC(MIC=2-4 µg/ml); Escherichia coli ATCC 25922 del-waaE(MIC=4 µg/ml); Escherichia coli ATCC 25922 del-waaF(MIC=2-4 µg/ml); Escherichia coli ATCC 25922 del-waaG(MIC=4 µg/ml); Escherichia coli JW3596(MIC=4 µg/ml); Escherichia coli JW1667(MIC=2-4 µg/ml ).
- [Ref.29852015]Gram-negative bacterial:Aeromonas salmonicida ATCC 33658(MIC=4 µg/ml);
- Gram-positive bacterial:Lactococcus lactis IL1403(MIC=1-2 µg/ml).
-
Hemolytic Activity
-
- [Ref.10768969]non-hemolysis against Human erythrocytes up to 500 µg/ml.
- [Ref.26656394]1% hemolysis against Horse erythrocytes at 64 µg/ml.
-
Cytotoxicity
-
- Not found
-
Binding Target
- liposomes
Structure Information
-
Linear/Cyclic
- Linear
-
N-terminal Modification
- Free
-
C-terminal Modification
- Free
-
Nonterminal Modifications and Unusual Amino Acids
- None
-
Stereochemistry
- L
-
Structure
- Alpha helix
-
Structure Description
- The peptide adopted an appreciable α-helical content in the presence of the organic cosolvent 40% trifluoroethanol and in the presence of 0.1% lipopolysaccharide.
-
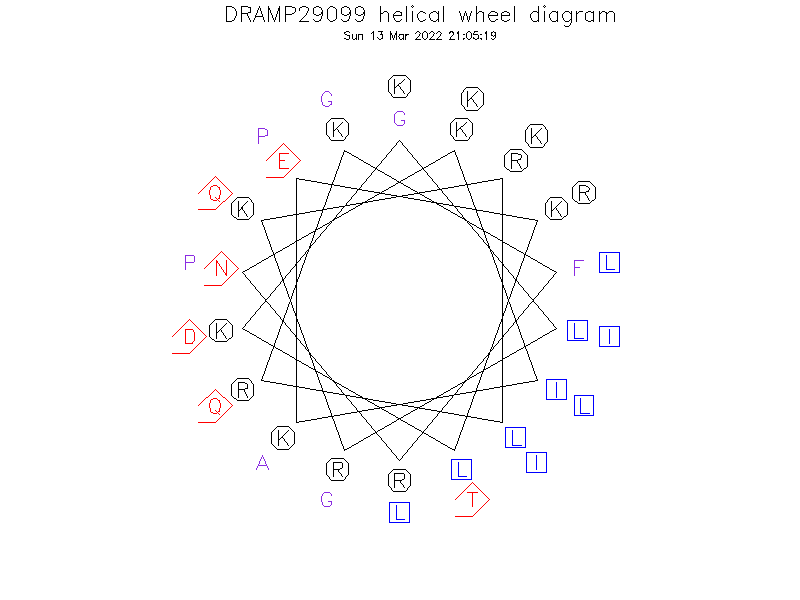
Helical Wheel Diagram
-
PDB ID
- 1LYP
-
Predicted Structure
- There is no predicted structure for DRAMP29099.
Physicochemical Information
-
Formula
- C202H356N64O47
Absent Amino Acids
- CHMSVW
Common Amino Acids
- K
Mass
- 4433.45
PI
- 11.54
Basic Residues
- 14
Acidic Residues
- 2
Hydrophobic Residues
- 11
Net Charge
- +12
-
Boman Index
- -10862
Hydrophobicity
- -1.097
Aliphatic Index
- 97.57
Half Life
-
- Mammalian:30 hour
- Yeast:>20 hour
- E.coli:>10 hour
Extinction Coefficient Cystines
- 1490
Absorbance 280nm
- 41.39
Polar Residues
- 6
DRAMP29099
Comments Information
Function
- Antibacterial activity against Gram-positive bacteria and Gram-negative bacteria.CAP18 were active against P. aeruginosa, E. coli, S. aureus, and MRSA.
Literature Information
- ·Literature 1
-
Title
- Cathelicidin peptides inhibit multiply antibiotic-resistant pathogens from patients with cystic fibrosis.
-
Pubmed ID
- 11557478
-
Reference
- Antimicrob Agents Chemother. 2001 Oct;45(10):2838-2844.
-
Author
- Saiman L, Tabibi S, Starner TD, San Gabriel P, Winokur PL, Jia HP, McCray PB Jr, Tack BF.
- ·Literature 2
-
Title
- Bactericidal activity of mammalian cathelicidin-derived peptides.
-
Pubmed ID
- 10768969
-
Reference
- Infect Immun. 2000 May;68(5):2748-2755.
-
Author
- Travis SM, Anderson NN, Forsyth WR, Espiritu C, Conway BD, Greenberg EP, McCray PB Jr, Lehrer RI, Welsh MJ, Tack BF.
- ·Literature 3
-
Title
- Comparative Evaluation of the Antimicrobial Activity of Different Antimicrobial Peptides against a Range of Pathogenic Bacteria.
-
Pubmed ID
- 26656394
-
Reference
- PLoS One. 2015 Dec 11;10(12):e0144611.
-
Author
- Ebbensgaard A, Mordhorst H, Overgaard MT, Nielsen CG, Aarestrup FM, Hansen EB.
- ·Literature 4
-
Title
- The Role of Outer Membrane Proteins and Lipopolysaccharides for the Sensitivity of Escherichia coli to Antimicrobial Peptides.
-
Pubmed ID
- 30245684
-
Reference
- Front Microbiol. 2018 Sep 7;9:2153.
-
Author
- Ebbensgaard A, Mordhorst H, Aarestrup FM, Hansen EB.
- ·Literature 5
-
Title
- Dissection of the antimicrobial and hemolytic activity of Cap18: Generation of Cap18 derivatives with enhanced specificity.
-
Pubmed ID
- 29852015
-
Reference
- PLoS One. 2018 May 31;13(5):e0197742.
-
Author
- Ebbensgaard A, Mordhorst H, Overgaard MT, Aarestrup FM, Hansen EB.