What is DRAMP Database?
DRAMP database is an information portal to biological active peptides. Peptides in this database come from three sources : Public databases (Swiss-Prot, PDB, PubMed), Clinical antimicrobial peptides (preclinical and clinical) and Patents.
Why DRAMP Database is created?
Antimicrobial peptides (AMPs) are widely distributed in nature and play a fundamental role in the innate immune response against pathogens. They have been treated as a novel therapeutic approach to combat bacterial infections in the presence of growing antibiotics resistance. For the convenience of researches on AMPs, a variety of useful antimicrobial peptide databases have been established, but many of them are at the level of specific classes or limited collection of AMPs or in lack of analytic tools. Considering this, we developed a comprehensive high-quality, user-friendly data repository of antimicrobial peptides (DRAMP).Entries in the database have detailed annotations, especially detailed antimicrobial activity data (shown as target organism with MIC value) and cytotoxicity data (shown as Measurement of Hemolytic Activity, MHC).The database comes with easy-to-operate browsing as well as searching with sorting and filtering functionalities. Besides, several useful sequence analysis tools are provided, including similarity search, sequence alignment and Conserved Domain Search (CD-Search). Compared with the current databases, DRAMP harbors extensive entries with biological activity information and holds a dataset of peptides in clinical development.
What is in DRAMP Database?
The current version of DRAMP holds 29946 antimicrobial sequences. Information related to peptides in DRAMP can be divided into seven parts:
- General information: DRAMP ID, Sequence (mature peptide), Sequence Length (all <=100), Peptide name/class, Gene, Source, Family, Swiss-Prot ID (for future information).
- Activity information: Biological Activity (e.g. Antibacterial), Target Organism (with detailed activity value), Hemolytic Activity, Cytotoxicity, Binding Targets (if known).
·The relationship map of classifications by activity in DRAMP. In the red box is the main classifications.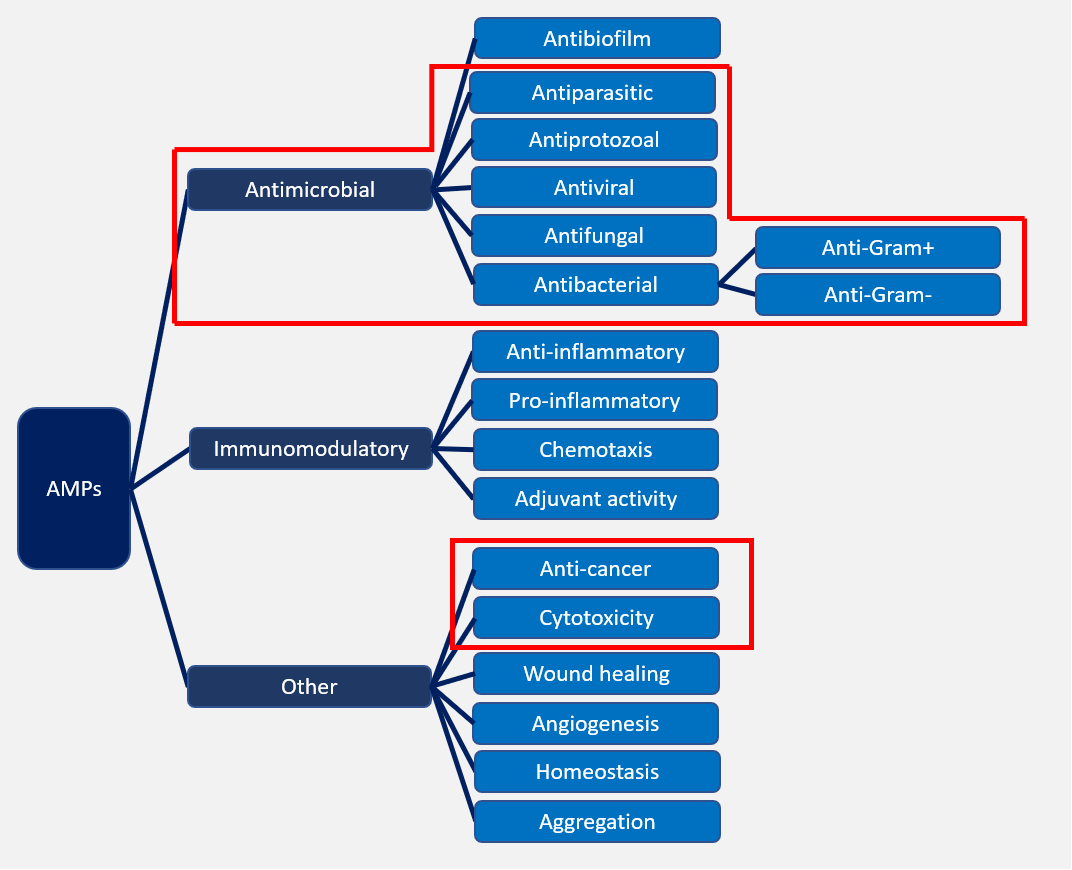
- Stability information: Serum or Protease Type, Stability Data, Assay.
- Structure information: Linear/Cyclic, N-terminal Modification, C-terminal Modification, Non-terminal modification, Stereochemistry, Structure (second structure), Structure Description (structure activity relationship), PDB ID (for future information), Predicted Structure.
- Physicochemical information: Length, Molecular weight, Theoretical pI, Amino acid composition, Net charge, Formula, Extinction coefficient, Estimated half-life (mammalian, yeast and E. coli), Instability index, Aliphatic index, Grand average of hydropathicity (GRAVY).
- Comments: Contain Function, Mode of action (MOA), Post-translational modification (PTM), and Toxicity (Hemolytic or Cytotoxicity), et al.
- Literature Information: Pubmed ID, Reference, Author and Title.
Disclaimer:
The Website is provided “as is” and the DRAMP hereby disclaim all warranties of any kind, express or implied. The DRAMP does not make any warranty that the services will be free of errors. You understand that you download from, or otherwise obtain content or services through, the DRAMP at your own discretion and risk.
The following people have contributed to creating DRAMP Database:
- Heng Zheng, Associate Professor of Pharmaceutical Sciences, work in Biomedicine Technology Transfer of Technology Transfer Platform; Nanjing University National Demonstration Base for Innovation and Entrepreneurship; School of Life Science and Technology, CPU; Institute of Medical and Pharmaceutical Biotechnology, Jiangsu Industrial Technology Research Institutegy. Prof. Zheng received his Ph.D in 2000, in microbial and biochemical pharmacy, CPU, under the supervision of Prof. Wutong Wu. In 2005-2007, he finished a postdoctoral work at School of Pharmaceutical Sciences, Nagasaki University, Japan. Prof. Zheng’s research focuses on the research and development of antimicrobial drugs, especially on the antimicrobial peptides. He is an expert in the field of computer aided drug design, discovered several novel metallo-β-lactamase inhibitors, which searched as potential drug candidates for multiple-drug-resistant bacteria. He is also specialize in protein structure prediction, molecular docking, and molecular dynamics. He published more than 80 papers, obtained 3 Chinese invention patents in recent years.
- Hanmei Xu, Professor of Pharmacy, Director of Department of Marine Pharmacy, School of Life Science and Technology; Ms. HanmeiXu, professor of CPU, supervisor of PH.D. students, head of the department of marine pharmacy in the college of life science and biotechnology, head of the engineering research center of peptide drug discovery and development of CPU. She has the experience of innovative peptide drug research and development for a long time and got the clinical approval for Antiangiotide, which is a class 1.1 chemical new drug in China. Currently, she is mainly engaged in innovation peptide drug design and discovery research and immunotherapy for cancer. Several innovative peptides such as anti-tumor, anti-rheumatoid and pulmonary fibrosis are being studied in pre-clinical stage. In recent years, she and her group have been founded by several national project and enterprise financing, more than 70 articles have been published, 35 invention patents have been applied.
- Jiahuang Li graduated from Nanjing University of technology, and got Bachelor's Degree and PhD Degree. Her major is Bioengineering, Protein Science and Structural bioinformatics. In 2004, she entered the State Key Laboratory of Pharmaceutical Biotechnology at Nanjing University and joined Professor ZichunHua’s group to carry out post-doctoral research. Since 2009, Ms. Jiahuang Li worked on college of life science in Nanjing University. Dr. Jiahuang Li have carried out a series of studies on Protein structure and function, Drug design and target screening, Molecular basis of genetic diseases and Tumor-targeting gene therapy. Dr. Li have published over a dozen SCI papers and got National Natural Science Foundation of China and the Jiangsu Provincial Nature Science Foundation.
- Xingzhen Lao, Associate Professor of Pharmaceutical Sciences,School of Life Science and Technology, CPU; Dr. Lao studied microbial and biochemical pharmacy at CPU and received her Ph.D. with Xiangdong Gao in 2012. Her major interests include peptide-receptor interaction, protein or peptide structure-functional analysis and peptide drugsdevelopment.
- Jian Sun graduated from CPU under the supervision of Associate Prof. Heng Zheng and has been working extensively in antimicrobial peptides.
- Linlin Fan graduated from CPU under the supervision of Associate Prof. Heng Zheng. Her major interests are in the field of antibacterial peptides and drug discovery.
- Shicai Liu graduated from CPU under the supervision of Associate Prof. Heng Zheng and has been working in computer aided drug design.
- Xinyue Kang graduated from CPU under the supervision of Associate Prof. Heng Zheng. She actively pursues the antimicrobial peptides research and computer aided drug design.
- Guobang Shi graduated from CPU under the supervision of Associate Prof. Heng Zheng. He actively pursues the antimicrobial peptides research and computer aided drug design.
- Yanchao Liu is studying Bioinformatics at CPU for the second year, under the supervision of Associate Prof. Heng Zheng. Now she is responsible for the daily update and maintenance and this is her email 2057850020@qq.com.
- Meifeng Zhou(Bachelor)
- Jie Zhou(PhD)








